Dimension reduction
Data dimension reduction method
feature selection
Redundancy: some features are highly correlated, affecting performance Noise: some characteristics have influence on the success of prediction 
Feature selection API
sklearn.feature_selection. VarianceThreshold By default, features with variance of 0 are deleted, and features with low variance are deleted. The threshold value is generally 0-10 (uncertain, depending on the actual situation) Feature selection code
#Feature Engineering - Feature Selection for Data Dimension Reduction from sklearn.feature_selection import VarianceThreshold def var(): """ Feature selection - delete low variance feature: return: None """ #Instantiation threshold is equal to the range of local difference. Here, all features with variance of 0 are deleted var = VarianceThreshold(threshold=0.0) data = var.fit_transform([[0, 2, 0, 3], [0, 1, 4, 3], [0, 1, 1, 3]]) print(data) return None if __name__ == "__main__": var() Operation results
[[2 0] [1 4] [1 1]] 
Principal component analysis (PCA)
There are not many application scenarios, and a large number of features will be used sklearn.decomposition N_components: decimal (usually 90%~95%), integer: (reduced number of features) 
code
#Feature Engineering - Principal Component Analysis (PCA) for Data Dimension Reduction from sklearn.decomposition import PCA def pca(): """ Principal component analysis for feature dimension reduction: return: None """ pca = PCA(n_components=0.9) data = pca.fit_transform([[0, 2, 0, 3], [0, 1, 4, 3], [0, 1, 1, 3]]) print(data) return None if __name__ == "__main__": pca() Operation results
[[-1.76504522] [ 2.35339362] [-0.58834841]] Principal component analysis case
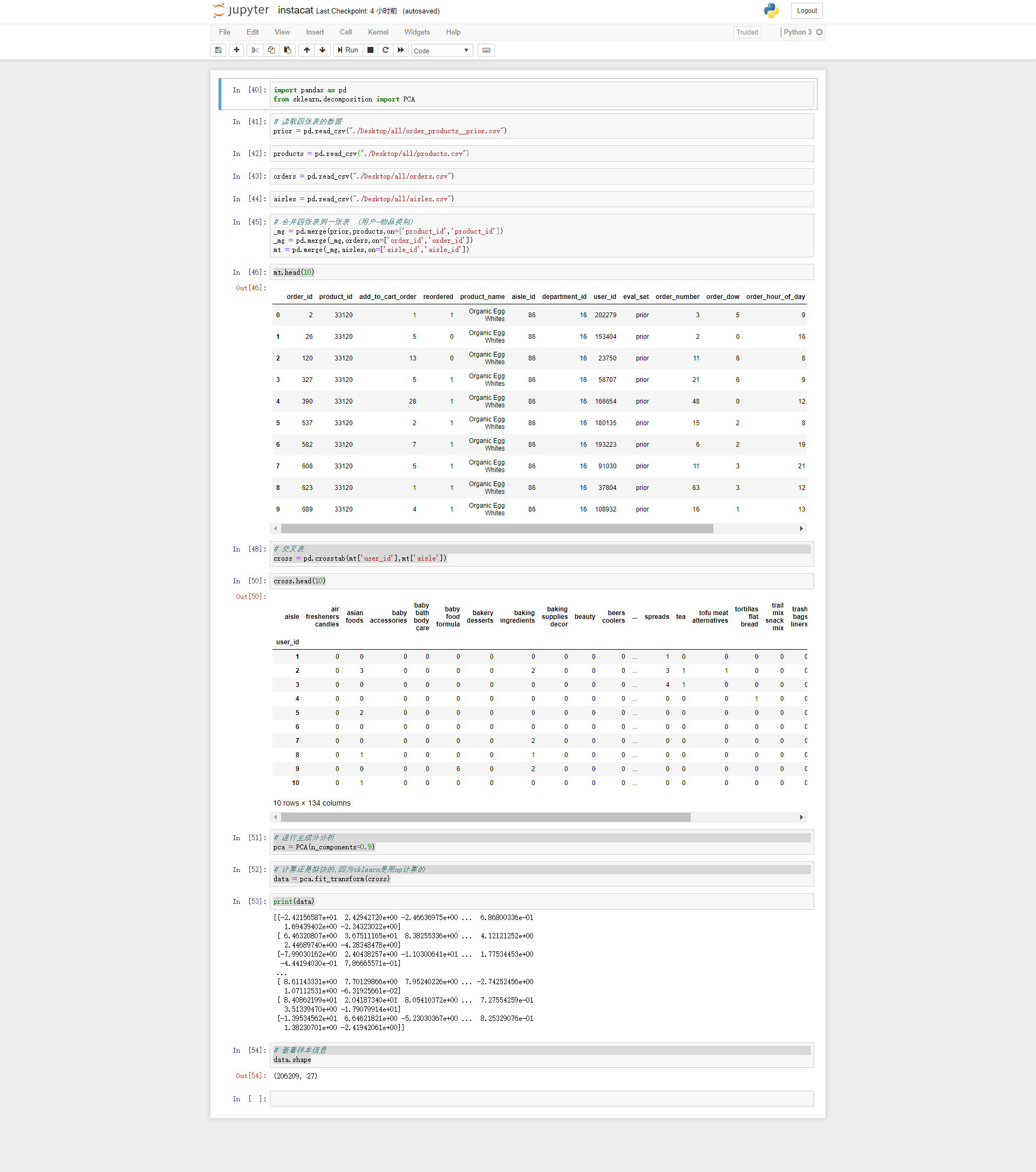
import pandas as pd from sklearn.decomposition import PCA #Read data from four tables prior = pd.read_csv("./Desktop/all/order_products__prior.csv") products = pd.read_csv("./Desktop/all/products.csv") orders = pd.read_csv("./Desktop/all/orders.csv") aisles = pd.read_csv("./Desktop/all/aisles.csv") #Consolidate four tables into one table (user item category) _mg = pd.merge(prior,products,on=['product_id','product_id']) _mg = pd.merge(_mg,orders,on=['order_id','order_id']) mt = pd.merge(_mg,aisles,on=['aisle_id','aisle_id']) mt.head(10) #Crosstab cross = pd.crosstab(mt['user_id'],mt['aisle']) cross.head(10) #Conduct principal component analysis pca = PCA(n_components=0.9) #The calculation is fast because sklearn is calculated with np data = pca.fit_transform(cross) print(data) #View sample information data.shape Operation results