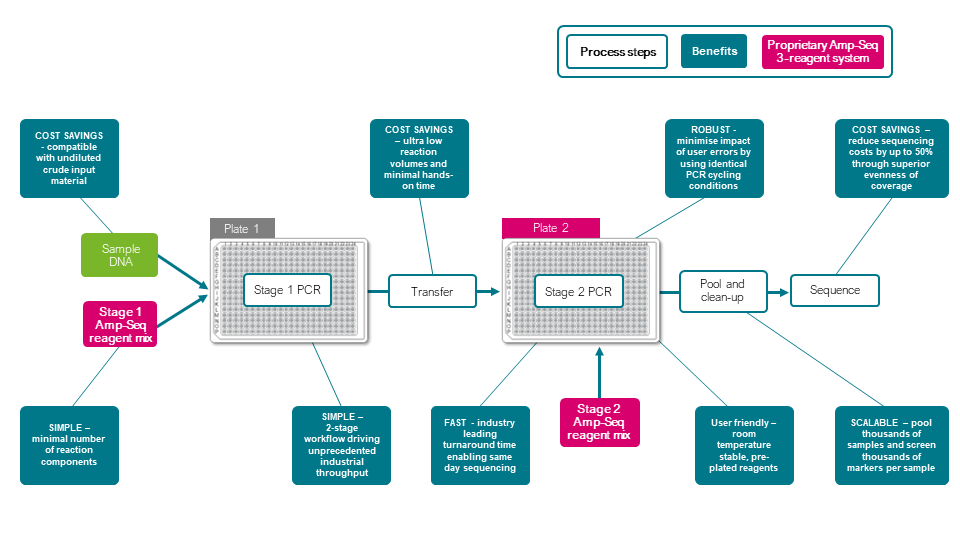
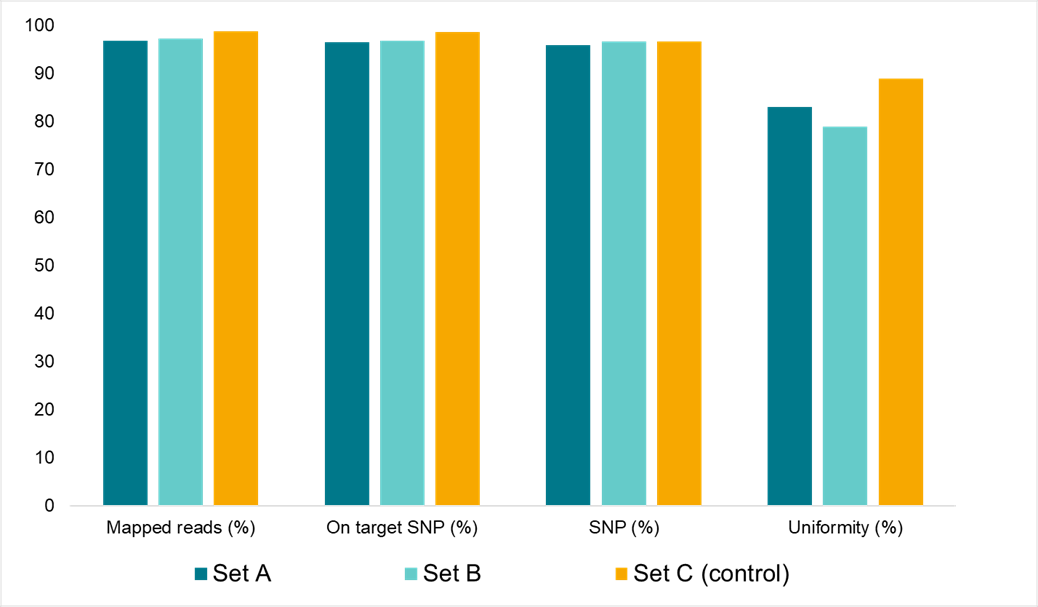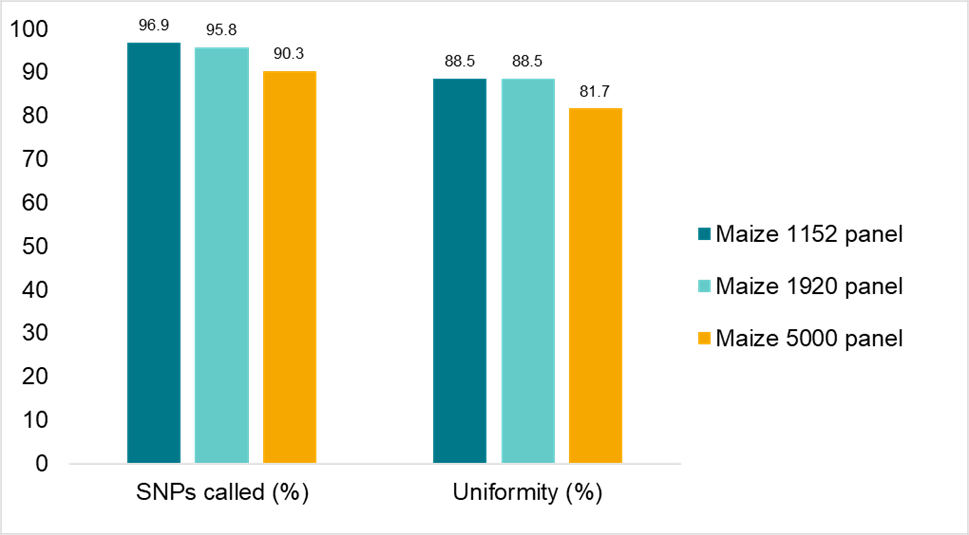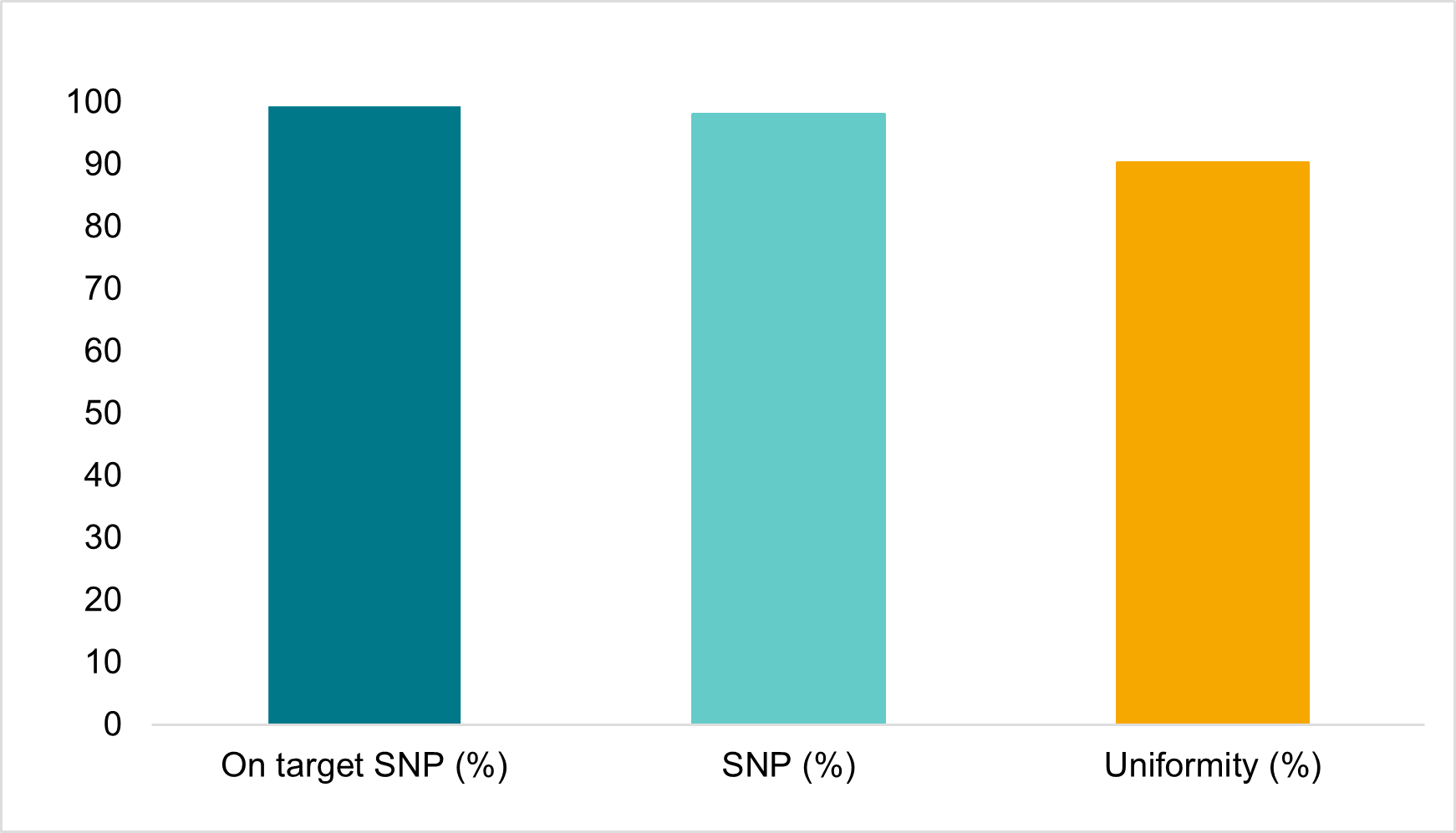质量,基本和性能
预应力试验,预应力试验和性能试验download the application note或,或start your custom evaluation。
质量控制
LGC has over180years of history as arecognised high-quality company and assay-to-assay consistency is established with the same rigorous quality controls。The assay reagents are optimised to achieve a high percentage of calls from the SNP panel design and a high SNP depth uniformity*for sample-to-sample data。All reagents are subjected to functional assays to establish high standards。锅炉炉膛安装和炉膛安装
Reagents are optimised during co-development of the targeting oligo library to be robust and suitable forgenerating high quality,uniform data across a range of sample input types。Newly designed primer pair pools are quality checked for optimal DNA input and optimal primer concentration to complete the robustness of the Amp-Seq assay。
*SNP depth unifor Amp-Seq is calculated as the percentage of SNPs with a depth above20%of the mean depth for that sample。
Assay stability
High-throughput automation of a large set of sample DNA requires that,at some stages in the protocol,384-well plates of reactions must sit at room temperature for several hours。
数据in figure2demonstrates that the reagents for the Amp-Seq assay are very stable at room temperature。
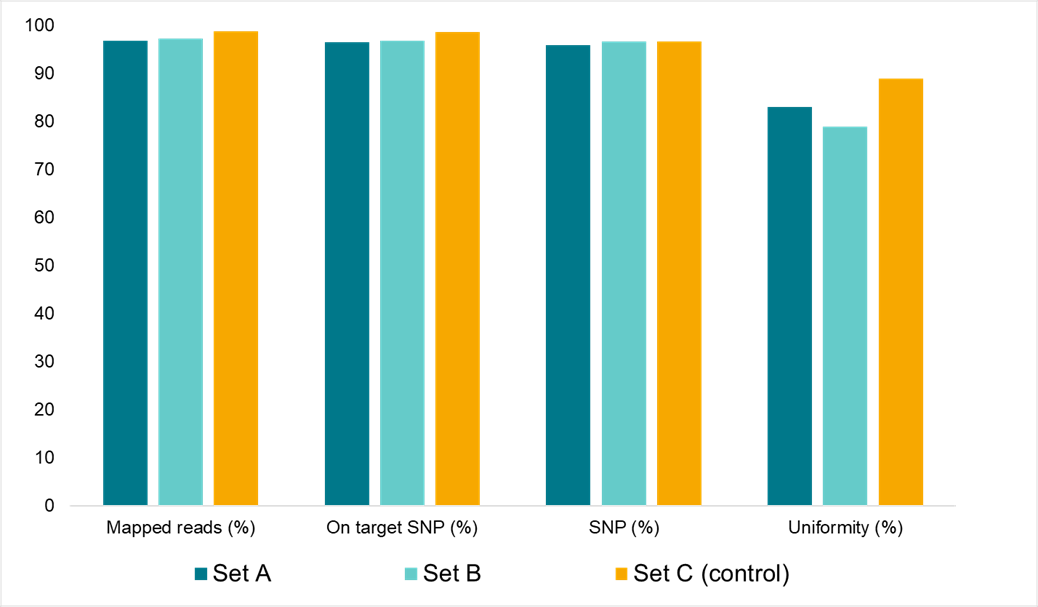
Figure2。Amp-Seq assay stability at room temperature。Sequencing data quality characteristics for soy samples processed through the workflow at different rates。设置;reaction mix was incubated for20hours at room temperature after stage1。Set B:reaction mix was incubated for 20hours at room temperature after stage1and after stage2。设置;control,all reactions were processed immediately with no room temperature hold for during either stage1 or stage2。
The stability of the Amp-Seq reagents allows for flexibility in running this assay at high-throughput due to the convenient handling of reagents。This simplification of the workflow contributes to the reduced turnaround times offered by Amp-Seq。
目标oligo library design service
Library design is performed in collaboration between the Biosearch Technologies team and the customer。目标规划contact your sales representative。
Software pipelines are used to design specific primer pools and for data analysis。The design software is rigorously tested to ensure that minimal primer interactions will occur,and that background signals are reduced to alvelthat does not significantly impact on-target mapping of amplicon reads。This level of stringency ensures that sequencing runs are efficient and very little sequence is wasted。Very high first pass design success rates are typically achieved,reducing the total turnaround time required for targeting oligo library design。
BiosearchCaller data analysis pipeline
The BiosearchCaller data analysis pipeline is available for installation and setup in your laboratory。It is designed to detect variants ensuring high variant calling accuracy and high genotype call concordance of diverse sample data when compared to other genotyping technologies。
协议自动化
数据in figure3displays the suitability of Amp-Seq for automation。一致性High performance was shown across384replicates of soy DNA,with a SNP call rate of over98%and a SNP depth uniformity of amplicon coverage of over90%on average。The flexibility to automate the workflow without impacting data quality further contributes to the reductions in turnaround times offered by Amp-Seq。
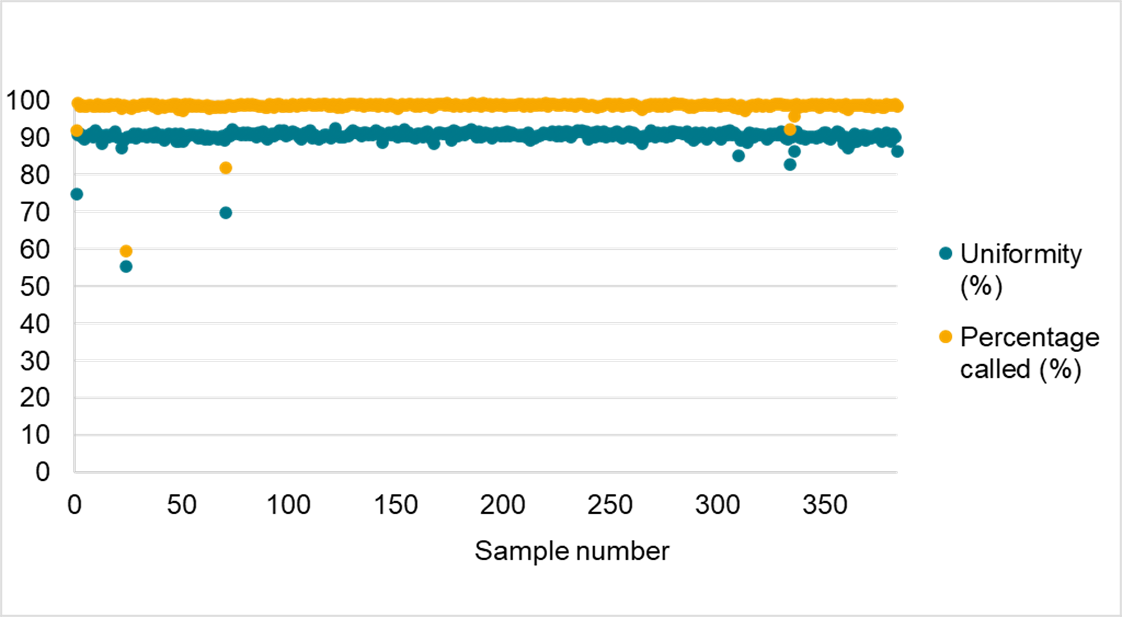
Figure3。Automation of the Amp-Seq assay。Performance of the soy1000+panel illustrates consistently high performance of the Amp-Seq assay when the protocol was automated using a Biomek liquid handler and dried index plates were used。
道路宽度-道路宽度
The robustness of the Amp-Seq assay was assessed with genomic DNA from several crops,including canola,maize,sorghum,soy,strawberry and wheat,using SNP panels of different sizes。Figure4illustrates data from maize panels of115521920and5000 SNPs that clearly demonstrates the flexibility and robustness of the assay。
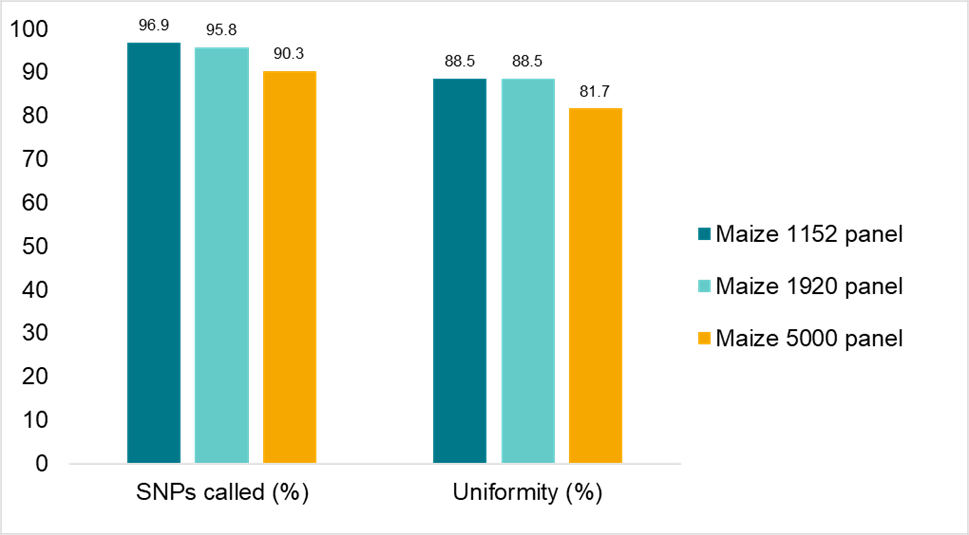
Figure4。Amp-Seq assay robustness across a range of panel sizes。MaizeB73 DNA was assayed with panels of 115521920and5000 primer pairs。Sequence data was downsampled to an average of 130.3131.1and115.7reads respectively。All panels passed our specification of 90%for percentage of SNPs called and 80%for SNP depth uniformity of amplicon reads。
Robustness–compatible with a variety of sample types-and DNA preparation methods
The robustness of the assay was also demonstrated using crude extracts from maize leaftissue as input as shown in figure5。This compatibility with undiluted crude input material allows for increased throughput and turnaround times by reducing upfront labour and logistics costs。
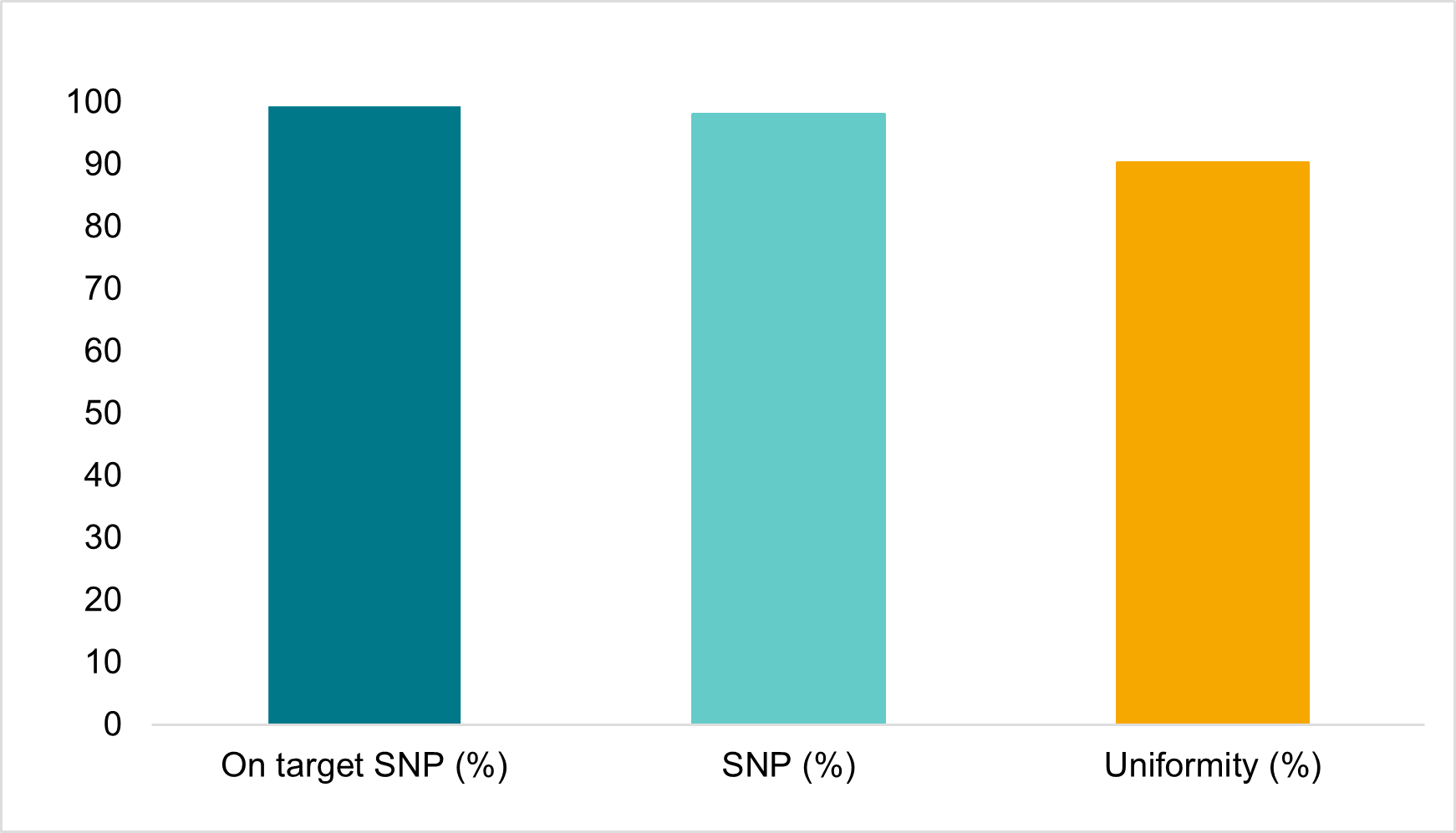
Figure5。Amp-Seq results for crudely extracted,undiluted sample material。Crude,undiluted maize leaf punch extract was used in the Amp-Seq protocol with a Maize1152SNP panel。The figure shows sequence analysis specifications for:i)percentage of reads that mapped to the correct sequence region(On target SNP%);ii)Percent of the 1152SNPs that were detected(SNP%)and iii)SNP depth uniformity(%)of mapped reads for the 1152SNPs at an average depth of 134reads。Quality of data will depend on the exact method of extraction。