基础,基础
The rapid and nearly unrestricted global spread of coronavirus disease(COVID-19)has resulted in the evolution of various mutants of severe acute respiratory syndrome coronavirus2(SARS-CoV-2)。With vaccines being the principal effective modality to curtail the pandemic,it is crucial to use them effectively and prepare for arise in the number of immune-escape mutants that can evolve due to the selection pressure exerted。Based on their clinical and epidemiological significance,the World Health Organization(WHO)has identified variant of concern(Alpha,Beta,Gamma,and Delta),variant of interest(Lambda and Mu),and variant s under monitoring(1个)。Although the Delta strain is the principal mutant responsible for the majority of the infections currently,variants with a few more amino acid substitutions in the Delta spike are emerging。
Previous studies have shown that mRNA vaccines such as BNT162b2 and mRNA-1273 confer robust保护against SARS-CoV-2(2)。However,several recent reports have shown that antibody titers drop markedly after6-8 months of vaccine administration(3–6个)。However,there has been no temporal and comprehensive study of neutralizing activities against the increasing number of delta derivatives。
Several human monoclonal antibodies have been used for the treatment of COVID-19,which contribute to the reduction of viral load and symptoms(7单击功能区上,8个)。However,some mutants have been shown to be resistant to these therapeutic antibodies,and the neutralizing capacity of the antibodies is greatly reduced(9单击功能区上,10)。
SARS-CoV-2neutralizing antibodies in sera within3h(11单击功能区上,12)。Therefore,by using hiVNT,we aimed to evaluate the efficacy of vaccine-derived neutralizing antibodies(nAbs)and therapeutic antibodies against the increasingly emerging recent variants。
矩阵和度量
项目和项目状态
Participants were recruited from among the medical staff of Yokohama City University Hospital in March 2021。Written informed consent was obtained from all the participants。Blood samples were collected1week and 6months after the administration of the second dose of Pfizer/BioNTech mRNA vaccine。Until the assessment date,we collected 126one-week sera samples and 98six-month sera samples,and all the samples were used。19 samples with blinding to demographic characteristics and designated this set as“Pvac 19 sera panel.”Prior to the experiment,all samples were tested for antibodies against SARS-CoV-2 spike and nucleocapsid protein and were confirmed to be positive and negative,respectively(there was no previous/breakthrough infection)。Blinding was not deemed necessary because the experiments did not involve any subjective assessment。No sample size calculation was performed。The study was conducted in accordance with the Declaration of Helsinki。This study was approved by the Yokohama City University Certified Institutional Review Board(Reference No.B21030001)。
Spike Haplotype Analysis
A302486 full genomes extracted from human subjects were downloaded from GISAID(13单击功能区上,14)and the National Center for Biotechnology Information(NCBI)up to September232021。In total,2400159 genomes meta data quality criterion of a<200bp gap。After a pairwise sequence alignment was performed with respect to the reference genome,we checked forimproper alignments which induce artifactual frameshifts and removed such sequences from further analysis。Furthermore,we eliminated the hyper-variant samples with over500mutations。We did not observe any recurrent stop gain mutations in our analysis。Variant annotation was performed as described in our previous report(15)。Briefly,a SARS-CoV-2genome was first aligned in a pairwise manner against the NC_045512 reference genome using the Needleman-Wunsch algorithm(16)and differences from the reference genome were extracted as genome changes and subsequently annotated for the types of variants and for amino acid changes。A set of variants associated with amino acid changes in the spike protein were extracted for each genome。Such a set of variants was called the spike haplotype。Distinct spike haplotypes were identified from the entire set of genomes。Next,spike haplotypes were assigned to each genome,including the subset spike haplotypes。Therefore,a single genome could be classified into multiple spike haplotypes。For instance,a Delta variant spike haplotype consisting of T19R,256_258 delinsG,L452R,T478K,D614G,P681R,and D950N is also assigned to another haplotype group of T19R,L452R,T478K,D614G,P681R,and D950N,which is missing a256_258delinsG。After grouping,the number of immune-escape variants,as reported previously(第十七节:–20),as well as the momentum,a metric of how quickly the frequency of a haplotype is increasing,were evaluated to identify the best candidates for antibody neutralization experiments。
拉pid Neutralization Test(HiVNT)
hiVNT was performed as described previously(11单击功能区上,12)。Briefly,the target cells seeded in 96-well plates were inoculated with50μL of HiBiT-tagged virus-like particles(hiVLPs)containing diluted serum(1:20-1:43740dilution for the quantitative assay;1:20dilution for the qualitative assay)。Intracellular luciferase activity was measured at3h after inoculation。
道路道路道路设计的道路:
应注意的是,应力不足,应力不足,应力不足,应力不足,应力不足50.We calculated the hiVNT50value using the curve-fitting tool ImageJ(NIH)。为什嚒不做某事50,it was assigned a hiVNT50value of 10。Alternatively,cells were inoculated with50μL of hiVLPs containing diluted antibody(final concentration of 0.64-50000ng/mL for REGN-COV2and 0.32-250000ng/mL for LY-COV)。REGN-COV2and LY-COV were research grade and were obtained from ProteoGenix and Invivogen,respectively。不存在的不存在的不存在的不存在的不存在的不存在的不存在的不存在的不存在的不存在的不存在的不存在的不存在的不存在的不存在的不存在的不存在的不存在的不存在的不存在50.The antibodies were tested individually,and the cocktail was considered effective against the viral mutant if it was neutralized by at least one antibody in the cocktail。
结果,结果
Vaccine-Escape Variants
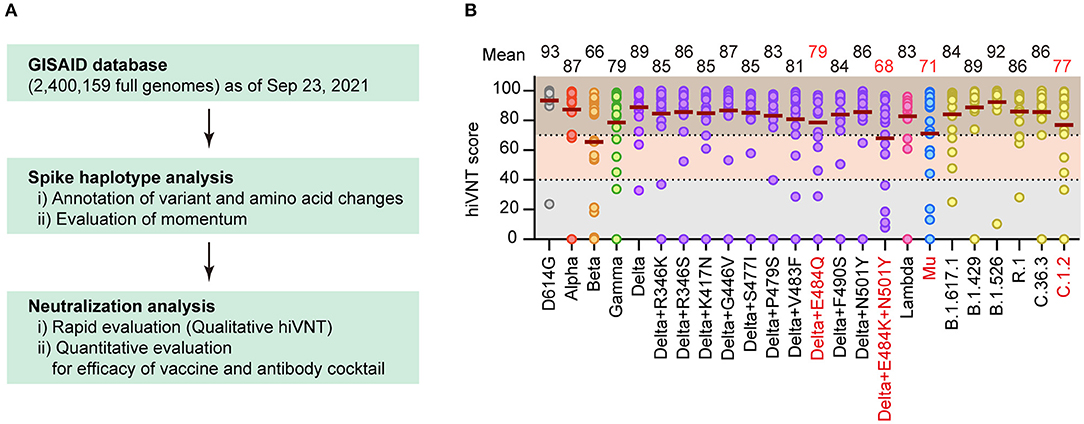
Of the 33002486 SARS-Cov-2full genomes downloaded from GISAID on September232021,we selected2400159 genomes that met the data quality criteria for the spike haplotype analysis。We identified12248distinct spike haplotypes(i.e.,sets of variants)with over10recurences from the whole genome setusing previously reported methods(15单击功能区上,16)。cases,the momentum,and immune escaping codons or mutations(第十七节:–20),we evaluated the number of immune-escape variants and the momentum to identify the best candidates for neutralization tests(火焰1A)。
To comprehensively identify the vaccine-escape strains,we performed a virus-like particle(VLP)-based rapid neutralization test(hiVNT)(11单击功能区上,12)on post-vaccination sera collected from individuals one week after administration of the second dose of the BNt162b2mRNA vaccine。In this study,a hiVNT score of 40 was set as the lower threshold,which is equivalent to 50%of the neutralizing titer against SARS-CoV-2pseudovirus(pvNT50)>50,并a hiVNT score of 70 was set as the higher threshold(equivalent to pvNT)50>200)(Supplementary Figure1)。预应力混凝土基础设施50insera of individuals with vaccine-breakthrough infections was approximately200(21)。Samples that fell below the lower threshold were considered to exhibit no neutralizing activity,those between the lower and higher threshold were considered to exhibit weak neutralizing activity,and those above the higher threshold were considered to exhibit stronutralizing activity。
A“Pvac19sera”panel(sera from19individuals collected one week after the second dose of Pfizer/BioNTech mRNA vaccine was administered)were used to determine the hiVNT score for each variant。The mean hiVNT score for most variants was approximately80,indicating that the vaccine could induce sufficient levels of neutralizing antibodies against these mutants as well。However,four variants,namely Beta and Delta derivatives(Delta+E484Q,Delta+E484K+N501Y),Mu,and C.1.2,showed relatively low hiVNT scores(火焰1B),suggesting that the neutralizing activity of post-vaccination sera against these variants might be weak。
SARS-CoV-2 Variants by Vaccine Sera and Therapeutic Antibodies
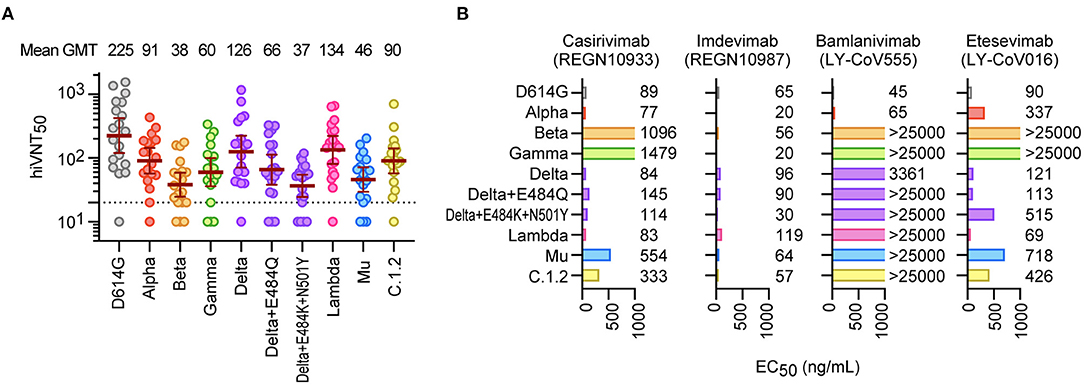
Next,we quantitatively evaluated the neutralizing activity against these variants。The serum dilution factor that inhibits VLP entry by half(hiVNT50)was assessed to demonstrate the neutralizing activity of the sera against these variants。The geometric mean titers(GMTs)were225 for D614G,38for Beta,and37 for Delta+E484K+N501Y(火焰2A),suggesting that the sera had6-fold reduced neutralization efficicacy against the Beta and Delta variants。However,the GMTs for all variants were above the effective threshold,suggesting that the vaccine-derived nAbs can neutralize the majority of variants tested。
We then evaluated the efficacy of the therapeutic antibodies(10单击功能区上,22),REGN10933(casirivimab),REGN10987(imdevimab),LY-COV555(bamlanivimab),and LY-COV016(etesevimab),against these mutants。因the casirivimab/imdevimab combination,all tested mutants were found to be neutralized by at least one of the two antibodies in the cocktail(火焰2B)。内contrast,bamlanivimab and etesevimab were less effective,especially against the Beta and Gamma strains(火焰2B)。Etesevimab was still effective against Delta,but the effect was reduced in Delta+E484K+N501Y。We further demonstrated that the Mu variant can also cause cell–cell fusion,similar to the Delta variant(Supplementary Figure2),which is highly likely to promote viral resistance to nAbs(23)。
Vaccine-Elicited Antibodies Against the Variants
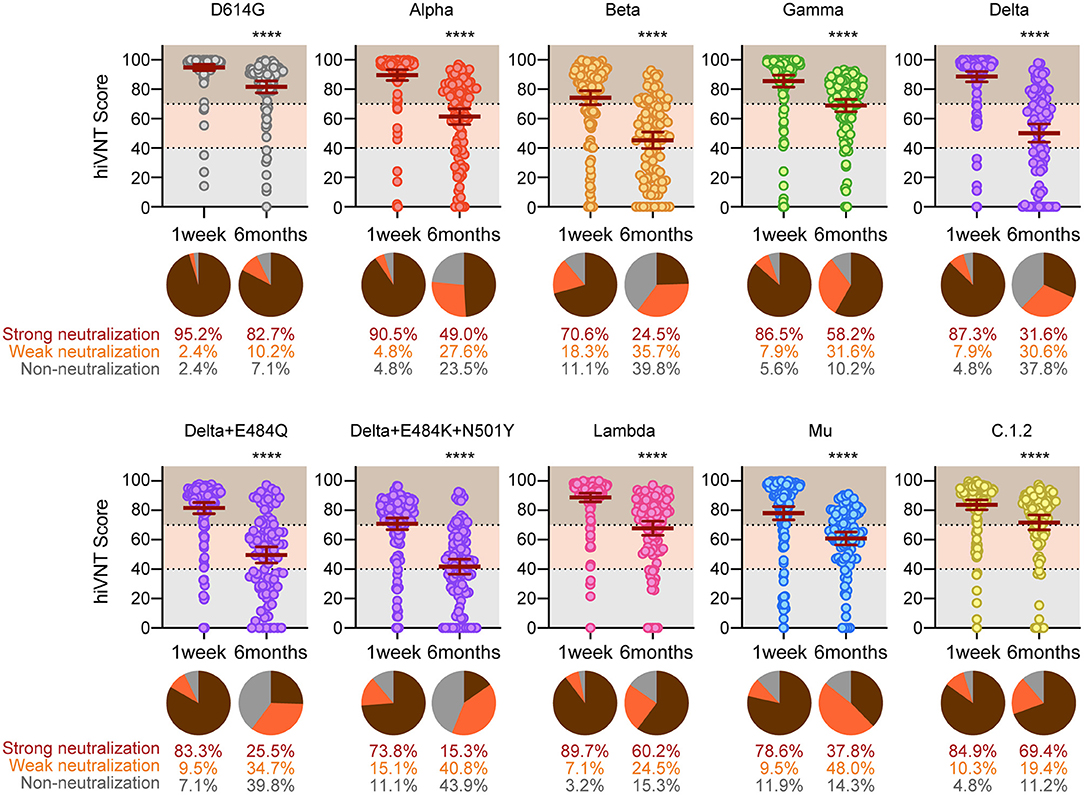
We recently reported that neutralizing antibody titers drop to 20%at6 months after vaccination(第二十四节:)。To examine the vaccine-elicited neutralizing antibody retention on a larger scale and over a longer period of time,we further increased the number of serum samples and compared the hiVNT scores of the variants at both1week(n=126)和6 months(n=98)post-vaccination。
At1week after vaccination,strong neutralization(hiVNT score>70)of all variant s was observed in most of the sera samples,ranging from the highest(95.2%)in D614G to the lowest in the Beta variant(70.6%)(Figure3)。Delta+E484K+N501Y and Mushowed a pattern similar to that of Beta,with73.8%and78.6%of the samples strongly neutralized,respectively。The proportion of sera samples that did not exhibit neutralizing activity was notably lower that of those exhibiting neutralizing activity for each variant。The highest occurrence of nAb escape(including weak and non-neutralizing activity,i.e.,hiVNT score<70)was noted with Beta(29.4%),followed by Delta+E484K+N501Y(26.2%)and Mu(21.4%)。This indicates that even immediately after two doses of mRNA vaccine,~20–30%of vaccinees may be at a risk of breakthrough infection of these variants。
Our results indicated that,at6months after vaccination,82.7%of the vaccinees exhibited strong neutralizing activity against the conventional strain。However,at6months after vaccination,strong neutralizing activity was significantly reduced against all mutant strains,ranging from the high est(60.2%)in the Lambda to the lowest in the Delta+E484K+N501Y variant(15.3%)(Figure3)。This result suggests that the strong neutralizing activity against SARS-Cov-2variants wane in 6months after vaccination,yet aweak neutralization is present。
电子气动装置,电子气动装置,电子气动装置,电子气动装置
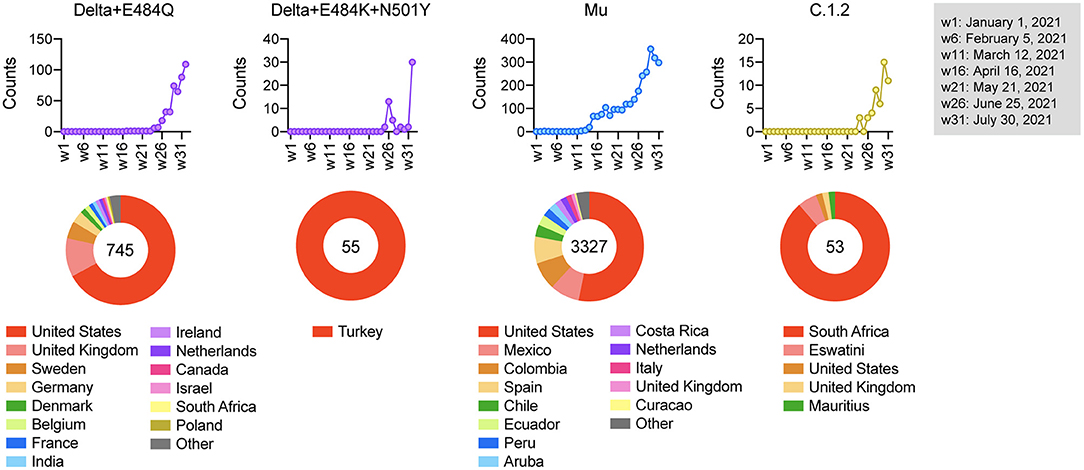
Finally,we examined the regions in which these strains of concern were mainly detected。Our epidemiological analysis demonstrated that the frequency of Delta+E484Q increased since week24of2021and the strain is still detected worldwide。Delta+E484K+N501Y was detected only in Turkey from week26,Muwas prevalent in South America from week14,and C.1.2was prevalent in South Africa frica from week26(火焰4)。The vaccination status in these countries is shown inSupplementary Figure4.We noticed that many of these haplotypes emerged before widespread vaccination,suggesting that vaccination might not be the likely cause of this emergence。Since vaccine-induced humoral immunity is less effective against these variants,their spread needs to be monitored carefully。
Discussion
In this study,by combining haplotype analysis and hiVNT,we identified immune evariant s that showed an increasing local trend。In addition,we tested the long-term efficacy of the Pfizer/BioNTech mRNA vaccine against these variants。
With the rise in emerging variants such as Delta derivatives,Mu,and C.1.2,concerns regarding the efficicacy of the currently available vaccines and antibody cocktail therapy have emerged。Our results show that the vaccine-derived nAbs and the antibody cocktail exhibit neutralization efficy against these variants。We observed this effect in the sera of vaccine recipients shortly after the administration of the second dose when the nAbs were considered to be at peak levels。
As vaccine-derived nAbs wane over time,follow-up studies are necessary to assess the persistence of nAbs against these variants。In fact,our analysis using sera6months after vaccination showed that the positive rate of nAb against the conventional strain was relatively maintained,while that against the mutant strains was markedly decreased。In particular,only about15-30%of vaccinees showed potent neutralizing activity against Delta,Delta+E484K+N501Y,and Mu strains。A comprehensive depiction of antibody prevalence by a hiVNT mutant panel not only allows for a rapid assessment of vaccine-elicited humoral immunity,but also highlights the need for booster vaccinations in areas where the mutant strains are prevalent。
Several reports have shown that after2-3 months of vaccination,the neutralizing activity on variant such as Delta strains is significantly lower than that of WT(25)。We have also shown a faster time-bound deterioration in neutralizing activity against the Delta strain(38%negative for neutralizing activity)than the WT strain。
Analysis of the therapeutic antibodies against the variants showed that imat imdevimab had high neutralizing activity against all the mutants tested,but casirivimab had reduced activity against Beta,Gamma,and Mu.These strains commonly include the E484K mutation,and this mutation is consimitrid with vimab,as previously indicated(10)。Unfortunately,bamlanivimab showed no neutralizing activity against the variants except Alpha,suggesting that it ineffective against the current prevalent strains。Etesevimab showed absolutely no neutralizing activity against Beta and Gamma,consistent with a previous report(9),and we found that this mAb was less effective against other mutant strains besides Delta and Lambda。TheN501Y mutation was common in the strains with reduced efficacy,suggesting that this mutation is a limitation of etesevimab。
Our results show that the Delta derivatives possess a high er vaccine escape than their parent Delta strain。Likewise,the Mu variant possesses a high er vaccine-escape ability than the Delta variant and also exhibits cell–cell fusion property like the latter。本发明提供了一种新的、新的、新的、新的、新的、新的、新的、新的、新的、新的、新的、新的、新的、新的、新的、新的、新的、新的、新的、新的、新的、新的、新的、新的26)。Therefore,such viral strains are more likely to evade humoral immunity。Hence,these variants could present a major challenge if either or both,or other immune escape mutants progresses to replace the Delta variant as the most predominantly transmitted variant。In the future,vaccines and therapeutic antibodies should be designed to address this problem。
数据可用性状态
The data analyzed in this study is subject to the following licenses/restrictions:The list of analyzed genomes from the GISAID's EpiFlu database is available from the corresponding author upon reasonable request。Requests to access these datasets should be directed to Kei Miyakawa,keim@yokohama-cu.ac.jp。
Ethics Statement
This study was approved by the Yokohama City University Certified Institutional Review Board(Reference No.B21030001)。The patients/participants provided their written informed consent to participate in this study。
Author Contributions
KM designed and performed the research,interpreted the data,and wrote the manuscript。SJ and RT interpreted the data and wrote the manuscript。YY performed the research and interpreted the data。TK designed and performed the spike haplotype analysis,interpreted the data,and wrote the manuscript。K interpreted the data。HK collected the specimens。AR directed the research,interpreted the data,and wrote the manuscript。All authors contributed to the article and approved the submitted version。
Funding
This study was supported by AMED grants(JP20he0522001and JP21fk0108104)to AR。
Conflict of Interest
YY is a current employee of Kanto Chemical Co.,Inc.TK,RT,and MK are current employees of IBM。
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as potential conflict of interest。
公共节点
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations,or those of the publisher,the editors and the reviewers。Any product that may be evaluated in this article,or claim that may be made by its manufacturer,is not guaranteed or endorsed by the publisher。
Acknowledgments
We acknowledge the support of the medical staff involved in this study。We thank Kenji Yoshihara,Kiho Tanaka,and Kazuo Horikawa for their technical assistance。We also gratefully acknowledge the authors and the originating and submitting laboratories providing sequences from the GISAID's EpiFlu™,™数据库和NCBI。
摘要材质
this article can be found online at:https://www.frontiersin.org/articles/10.3389/fmed.2022.811004/full#supplementary-material
参考,参考
1.WHO。Trackking SARS-COV-2variants。Washinton,DC:WHO(2020)。
Google Scholar
2.Walsh EE,Frenck RW Jr,Falsey AR,Kitchin N,Absalon J,Gurtman A,et al.Gruber,safety and immunogenicity of two RNA-based Covid-19vaccine candidates。N Engl J Med。(2020)383:2439–50。doi:10.01056/NEJMoa2027906
PubMed Abstract|CrossRef Full Text|Google Scholar
3.Collier AY,Yu J,McMahan K,Liu J,Chandrashekar A,Maron JS,et al.Differential kinetics of immune responses elicited by Covid-19vaccines。N Engl J Med。(2021)385:2010–2。doi:10.1056/NEJMc2115596
PubMed Abstract|CrossRef Full Text|Google Scholar
4.Pegu A,O'Connell SE,Schmidt SD,O'Dell S,Talana CA,Lai L,et al.Durability of mRNA-1273vaccine-induced antibodies against SARS-COV-2variants。Science。(2021)373:1372–7。doi:10.1126/science.abj4176
PubMed Abstract|CrossRef Full Text|Google Scholar
6.Naaber P,Tserel L,Kangro K,Sepp E,Jurjenson V,Adamson A,et al.Dynamics of antibody response to BNT162b2vaccine after six months:a longitudinal prospective study。Lancet Reg Health Eur。(2021)10:100208。doi:10.1016/j.lanepe.2021.100208
PubMed Abstract|CrossRef Full Text|Google Scholar
7.Weinreich DM,Sivapalasingam S,Norton T,Ali S,Gao H,Bhore R,et al.REGN-COV2,aneutralizing antibody cocktail,in outpatients with Covid-19。N Engl J Med。(2021)384:238-51。doi:10.1056/NEJMoa2035002
PubMed Abstract|CrossRef Full Text|Google Scholar
8.Chen P,Nirula A,Heller B,Gottlieb RL,Boscia J,Morris J,et al.SARS-CoV-2neutralizing antibody LY-CoV555in outpatients with Covid-19。N Engl J Med。(2021)384:229-37。doi:10.1056/NEJMoa2029849
PubMed Abstract|CrossRef Full Text|Google Scholar
10.Hoffmann M,Arora P,Gross R,Seidel A,Hornich BF,Hahn AS,et al.SARS-Cov-2variants B.1.351and P.1escape from neutralizing antibodies。中心,中心.(2021)184:2384–93.e12。doi:10.1016/j.cell.2021.03.036
PubMed Abstract|CrossRef Full Text|Google Scholar
11.Miyakawa K,Jeremiah SS,Ohtake N,Matsunaga S,Yamaoka Y,Nishi M,et al.Rapid quantitative screening assay for SARS-CoV-2neutralizing antibodies using HiBiT-tagged virus-like particles。J Mol Cell Biol。(2020)12:987–90。doi:10.1093/jmcb/mjaa047
PubMed Abstract|CrossRef Full Text|Google Scholar
12.Miyakawa K,Jeremiah SS,Kato H,Yamaoka Y,Go H,Yajima S,et al.Rapid detection of neutralizing antibodies to SARS-CoV-2variants in post-vaccination sera。J Mol Cell Biol。(2021)。doi:10.1101/2021.05.06.21256788。[Epub ahead of print]。
PubMed Abstract|CrossRef Full Text|Google Scholar
16.Needleman SB,Wunsch CD.A general method applicable to the search for similarities in the amino acid sequence of two proteins。J Mol Biol。(1970)48:443-53。doi:10.1016/0022-2836(70)90057-4
PubMed Abstract|CrossRef Full Text|Google Scholar
17.Wang R,Chen J,Gao K,Wei GW.Vaccine-escape and fast-growing mutations in the United Kingdom,the United States,Singapore,Spain,India,and other COVID-19-devastated countries。Genomics。(2021)113:2158–70。doi:10.1016/j.ygeno.2021.05.006
PubMed Abstract|CrossRef Full Text|Google Scholar
18.Liu Z,VanBlargan LA,Bloyet LM,Rothlauf PW,Chen RE,Stumpf S,et al.Identification of SARS-CoV-2spike mutations that attenuate monoclonal and serum antibody neutralization。Cell Host Microbe。(2021)29:477–88.e4。doi:10.1016/j.chom.2021.014
PubMed Abstract|CrossRef Full Text|Google Scholar
19.Harvey WT,Carabelli AM,Jackson B,Gupta RK,Thomson EC,Harrison EM,et al.SARS-CoV-2variants,spike mutations and immune escape。Nat Rev Microbiol。(2021)19:409-24。doi:10.1038/s41579-021-00573-0
PubMed Abstract|CrossRef Full Text|Google Scholar
20.Wang L,Zhou T,Zhang Y,Yang ES,Schramm CA,Shi W,et al.Ultrapotent antibodies against diverse and high ly transmissible SARS-CoV-2variants。Science。(2021)373:eabh1766。doi:10.1126/science.abh1766
PubMed Abstract|CrossRef Full Text|Google Scholar
22.Baum A,Fulton BO,WLoga E,Copin R,Pascal KE,Russo V,et al.Antibody cocktail to SARS-CoV-2spike protein prevents rapid mutational escape seen with individual antibodies。Science。(2020)369:1014-8。doi:10.1126/science.abd0831
PubMed Abstract|CrossRef Full Text|Google Scholar
24.Kato H,Miyakawa K,Ohtake N,Yamaoka Y,Yajima S,Yamazaki E,et al.Vaccine-induced humoral and cellular immunity against SARS-COV-2at6months post BNT162b2vaccination。medRxiv[2011.0.30.21265693]。(2021)。doi:10.1101/2021.10.30.21265693
CrossRef Full Text|Google Scholar
25.Planas D,Veyer D,Baidaliuk A,Staropoli I,Guivel-Benhassine F,Rajah MM,et al.Reduced sensitivity of SARS-CoV-2variant Delta to antibody neutralization。Nature。(2021)596:276-80。doi:10.1038/s41586-021-03777-9
PubMed Abstract|CrossRef Full Text|Google Scholar