This should be the simplest and most useful GSEA enrichment analysis tool in history
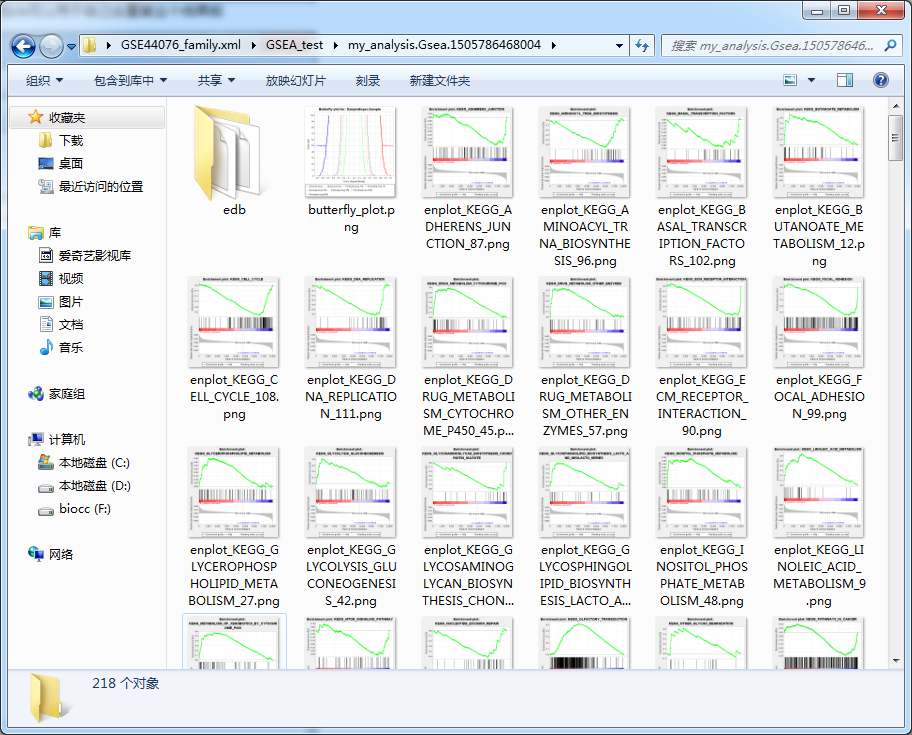
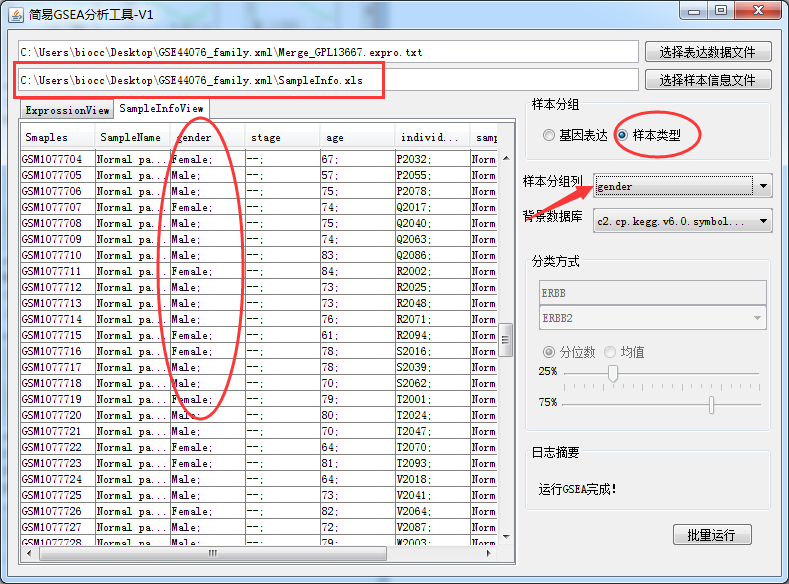
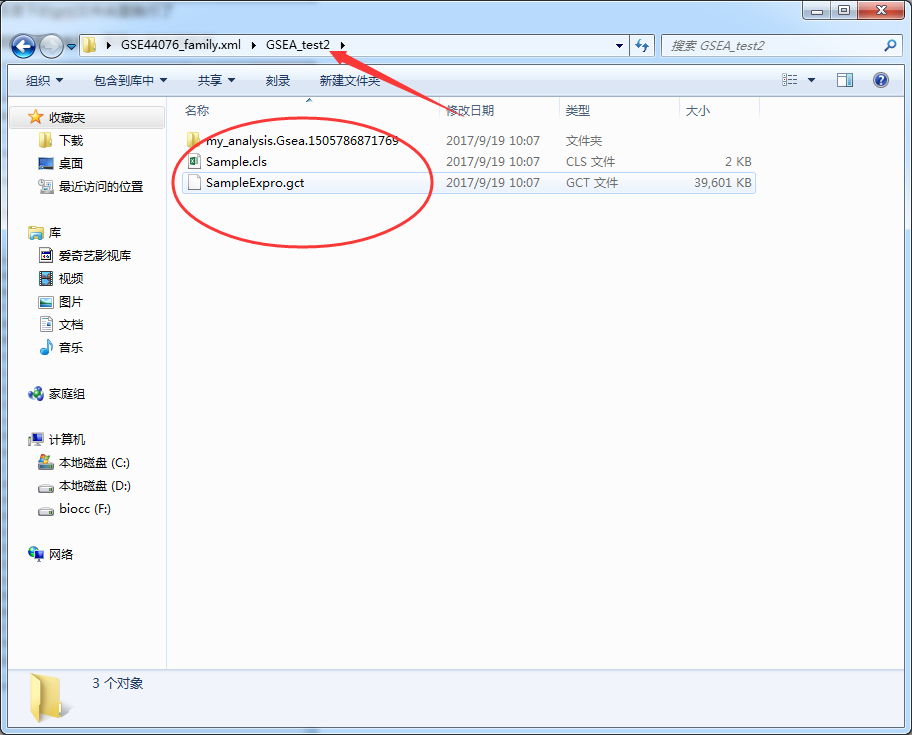
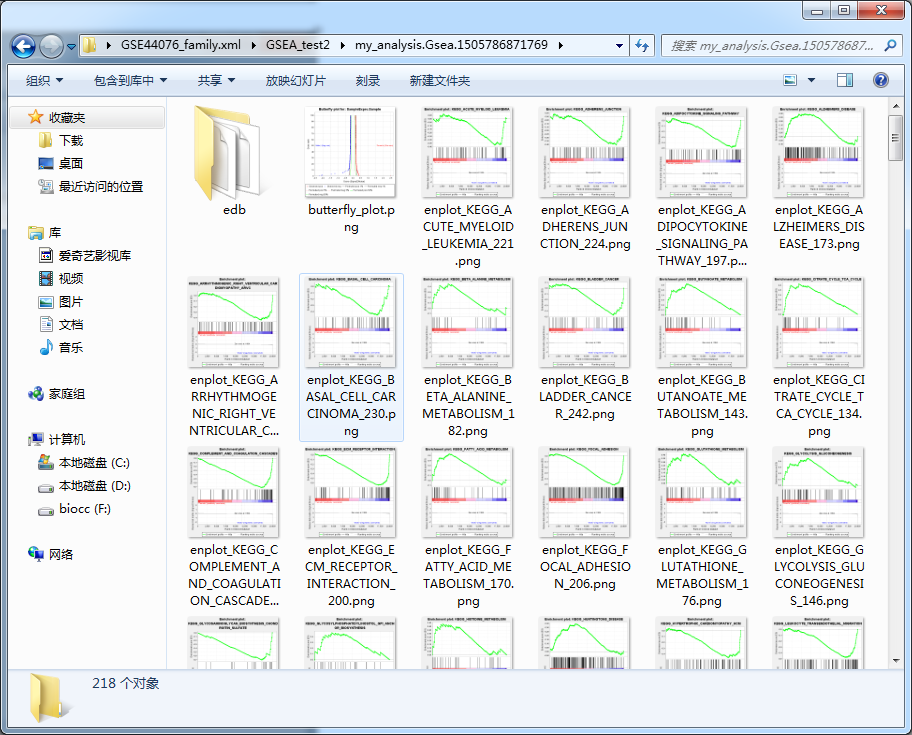
-
Published on 2017-09-19 10:12 -
Reading (57581)
Articles you may be interested in
-
GSEA simple analysis and visualization tool 3970 Browse -
Brief Introduction of GSEA Enrichment Analysis 15612 Browse -
Analysis of the principle of ssGSEA algorithm 30434 Browse -
GEO and TCGA database mining article ideas! 28079 Browse -
Five minutes of hands will teach you how to do GESA enrichment analysis 21532 Browse
Related issues
-
The GSEA gadget on the sangerbox webpage runs twice, using the same matrix data, grouping and geneset, but the results are different 1 Answer -
What's wrong with the non response of GSEA gadget points after batch running? 0 Answer -
What about the NES value, p value and FDR value after GSEA enrichment analysis in sangerbox analysis TCGA database 1 Answer -
How to choose the classification method of GSEA gadgets? What is the impact of this method 1 Answer -
No response in GSEA analysis (msigdb C2 data set is used) 1 Answer -
GSEA has issued many documents, and I don't know which is effective 1 Answer
56 comments
Writer list »
-

Zhu Rangfei 118 Articles -

grapefruit 91 Articles -

Liu Yongxin 64 Articles -

admin 57 Articles -

Shengxin Analysis Flow 55 Articles -

SXR 44 Articles -

Helen Zhang 31 Articles -

Shuanger 25 Articles
