Abstract
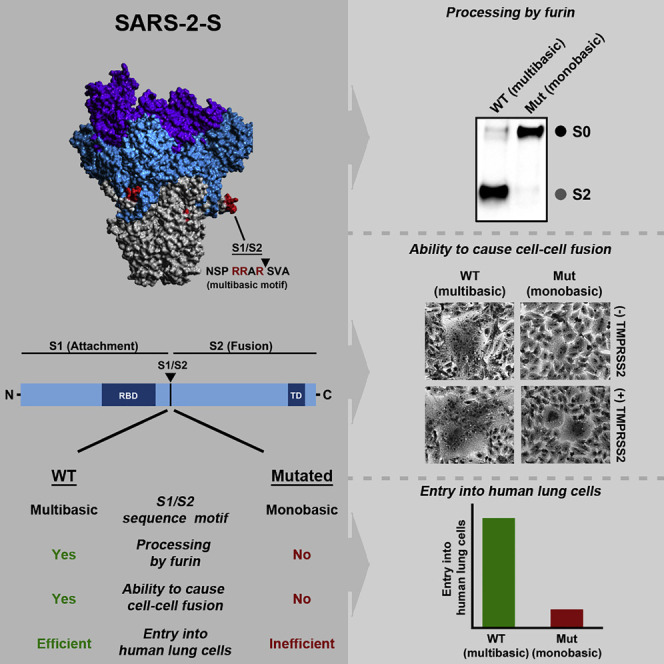
Graphical Abstract
Introduction
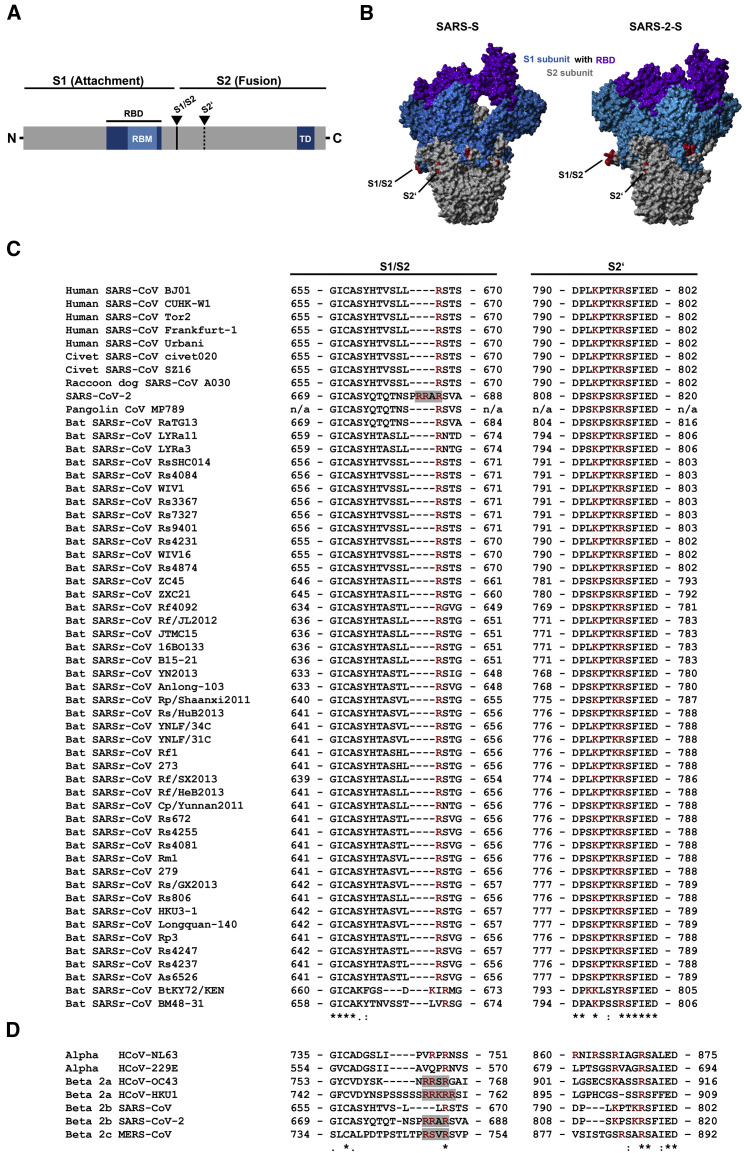
Figure 1.
Results
The Multibasic S1/S2 Site in the Spike Protein of SARS-CoV-2 Is Required for Efficient Proteolytic Cleavage of the Spike Protein
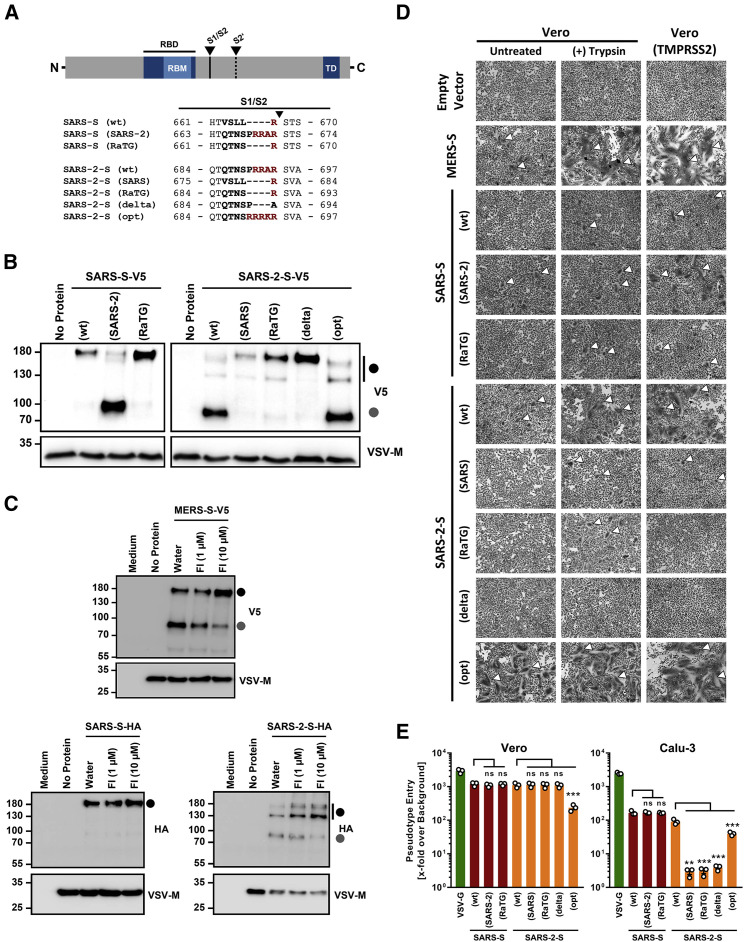
Figure 2.
Furin Cleaves the SARS-CoV-2 Spike Protein at the S1/S2 Site, and Cleavage Is Required for Efficient Cell-Cell Fusion
Cleavage of the SARS-CoV-2 Spike Protein at the S1/S2 Site Is Required for Viral Entry into Human Lung Cells
Discussion
Limitations of the Study
STAR ★ Methods
Key Resources Table
|
|
||
|
|
||
Resource Availability
Lead Contact
Materials Availability
Data and Code Availability
Method Details
Cell cultures
Plasmids
Preparation of pseudotyped particles and transduction experiments
Western blot analysis
Syncytium formation assay
Sequence analysis and protein models
Quantification and Statistical Analysis
Acknowledgments
Author Contributions
Declaration of Interest
References
-
Berger Rentsch M., Zimmer G. A vesicular stomatitis virus replicon-based bioassay for the rapid and sensitive determination of multi-species type I interferon. PLoS ONE. 2011; 6:e25858. doi: 10.1371/journal.pone.0025858. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ] -
Bertram S., Dijkman R., Habjan M., Heurich A., Gierer S., Glowacka I., Welsch K., Winkler M., Schneider H., Hofmann-Winkler H., et al. TMPRSS2 activates the human coronavirus 229E for cathepsin-independent host cell entry and is expressed in viral target cells in the respiratory epithelium. J. Virol. 2013; 87:6150–6160. doi: 10.1128/JVI.03372-12. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ] -
Brinkmann C., Hoffmann M., L ü bke A., Nehlmeier I., Kr ä mer-K ü hl A., Winkler M., P ö hlmann S. The glycoprotein of vesicular stomatitis virus promotes release of virus-like particles from tetherin-positive cells. PLoS ONE. 2017; 12:e0189073. doi: 10.1371/journal.pone.0189073. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ] -
Ge X.Y., Li J.L., Yang X.L., Chmura A.A., Zhu G., Epstein J.H., Mazet J.K., Hu B., Zhang W., Peng C., et al. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature. 2013; 503:535–538. doi: 10.1038/nature12711. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ] -
Gierer S., Bertram S., Kaup F., Wrensch F., Heurich A., Kr ä mer-K ü hl A., Welsch K., Winkler M., Meyer B., Drosten C., et al. The spike protein of the emerging betacoronavirus EMC uses a novel coronavirus receptor for entry, can be activated by TMPRSS2, and is targeted by neutralizing antibodies. J. Virol. 2013; 87:5502–5511. doi: 10.1128/JVI.00128-13. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ] -
Gierer S., M ü ller M.A., Heurich A., Ritz D., Springstein B.L., Karsten C.B., Schendzielorz A., Gnir ß K., Drosten C., P ö hlmann S. Inhibition of proprotein convertases abrogates processing of the middle eastern respiratory syndrome coronavirus spike protein in infected cells but does not reduce viral infectivity. J. Infect. Dis. 2015; 211:889–897. doi: 10.1093/infdis/jiu407. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ] -
Glowacka I., Bertram S., M ü ller M.A., Allen P., Soilleux E., Pfefferle S., Steffen I., Tsegaye T.S., He Y., Gnirss K., et al. Evidence that TMPRSS2 activates the severe acute respiratory syndrome coronavirus spike protein for membrane fusion and reduces viral control by the humoral immune response. J. Virol. 2011; 85:4122–4134. doi: 10.1128/JVI.02232-10. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ] -
Hoffmann M., Hofmann-Winkler H., P ö hlmann S. In: Activation of Viruses by Host Proteases. B ö ttcher-Friebertsh ä user E., Garten W., Klenk H., editors. Springer; 2018. Priming Time: How Cellular Proteases Arm Coronavirus Spike Proteins; pp. 71–98. [ Google Scholar ] -
Hoffmann M., M ü ller M.A., Drexler J.F., Glende J., Erdt M., G ü tzkow T., Losemann C., Binger T., Deng H., Schwegmann-We ß els C., et al. Differential sensitivity of bat cells to infection by enveloped RNA viruses: coronaviruses, paramyxoviruses, filoviruses, and influenza viruses. PLoS ONE. 2013; 8:e72942. doi: 10.1371/journal.pone.0072942. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ] -
Hoffmann M., Kleine-Weber H., Schroeder S., Kr ü ger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 2020; 181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ] -
Hulswit R.J., de Haan C.A., Bosch B.J. Coronavirus Spike Protein and Tropism Changes. Adv. Virus Res. 2016; 96:29–57. doi: 10.1016/bs.aivir.2016.08.004. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ] -
Iwata-Yoshikawa N., Okamura T., Shimizu Y., Hasegawa H., Takeda M., Nagata N. TMPRSS2 Contributes to Virus Spread and Immunopathology in the Airways of Murine Models after Coronavirus Infection. J. Virol. 2019; 93:93. doi: 10.1128/JVI.01815-18. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ] -
Kleine-Weber H., Elzayat M.T., Hoffmann M., P ö hlmann S. Functional analysis of potential cleavage sites in the MERS-coronavirus spike protein. Sci. Rep. 2018; 8:16597. doi: 10.1038/s41598-018-34859-w. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ] -
Kleine-Weber H., Elzayat M.T., Wang L., Graham B.S., M ü ller M.A., Drosten C., P ö hlmann S., Hoffmann M. Mutations in the Spike Protein of Middle East Respiratory Syndrome Coronavirus Transmitted in Korea Increase Resistance to Antibody-Mediated Neutralization. J. Virol. 2019; 93:93. doi: 10.1128/JVI.01381-18. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ] -
Krieger E., Vriend G. YASARA View - molecular graphics for all devices - from smartphones to workstations. Bioinformatics. 2014; 30:2981–2982. doi: 10.1093/bioinformatics/btu426. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ] -
Lam T.T., Shum M.H., Zhu H.C., Tong Y.G., Ni X.B., Liao Y.S., Wei W., Cheung W.Y., Li W.J., Li L.F., et al. Identifying SARS-CoV-2 related coronaviruses in Malayan pangolins. Nature. 2020 doi: 10.1038/s41586-020-2169-0. Published online March 26, 2020. [ DOI ] [ PubMed ] [ Google Scholar ] -
Li X., Zai J., Zhao Q., Nie Q., Li Y., Foley B.T., Chaillon A. Evolutionary history, potential intermediate animal host, and cross-species analyses of SARS-CoV-2. J. Med. Virol. 2020 doi: 10.1002/jmv.25731. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ] -
Luczo J.M., Stambas J., Durr P.A., Michalski W.P., Bingham J. Molecular pathogenesis of H5 highly pathogenic avian influenza: the role of the haemagglutinin cleavage site motif. Rev. Med. Virol. 2015; 25:406–430. doi: 10.1002/rmv.1846. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ] -
Madeira F., Park Y.M., Lee J., Buso N., Gur T., Madhusoodanan N., Basutkar P., Tivey A.R.N., Potter S.C., Finn R.D., Lopez R. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019; 47(W1):W636–W641. doi: 10.1093/nar/gkz268. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ] -
Matsuyama S., Nagata N., Shirato K., Kawase M., Takeda M., Taguchi F. Efficient activation of the severe acute respiratory syndrome coronavirus spike protein by the transmembrane protease TMPRSS2. J. Virol. 2010; 84:12658–12664. doi: 10.1128/JVI.01542-10. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ] -
Menachery V.D., Dinnon K.H., 3rd, Yount B.L., Jr., McAnarney E.T., Gralinski L.E., Hale A., Graham R.L., Scobey T., Anthony S.J., Wang L., et al. Trypsin Treatment Unlocks Barrier for Zoonotic Bat Coronaviruses Infection. J. Virol. 2020; 94:e01774-19. doi: 10.1128/JVI.01774-19. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ] -
Millet J.K., Whittaker G.R. Host cell entry of Middle East respiratory syndrome coronavirus after two-step, furin-mediated activation of the spike protein. Proc. Natl. Acad. Sci. USA. 2014; 111:15214–15219. doi: 10.1073/pnas.1407087111. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ] -
Millet J.K., Whittaker G.R. Physiological and molecular triggers for SARS-CoV membrane fusion and entry into host cells. Virology. 2018; 517:3–8. doi: 10.1016/j.virol.2017.12.015. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ] -
Park J.E., Li K., Barlan A., Fehr A.R., Perlman S., McCray P.B., Jr., Gallagher T. Proteolytic processing of Middle East respiratory syndrome coronavirus spikes expands virus tropism. Proc. Natl. Acad. Sci. USA. 2016; 113:12262–12267. doi: 10.1073/pnas.1608147113. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ] -
Roebroek A.J., Umans L., Pauli I.G., Robertson E.J., van Leuven F., Van de Ven W.J., Constam D.B. Failure of ventral closure and axial rotation in embryos lacking the proprotein convertase Furin. Development. 1998; 125:4863–4876. doi: 10.1242/dev.125.24.4863. [ DOI ] [ PubMed ] [ Google Scholar ] -
Sarac M.S., Cameron A., Lindberg I. The furin inhibitor hexa-D-arginine blocks the activation of Pseudomonas aeruginosa exotoxin A in vivo. Infect. Immun. 2002; 70:7136–7139. doi: 10.1128/IAI.70.12.7136-7139.2002. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ] -
Sarac M.S., Peinado J.R., Leppla S.H., Lindberg I. Protection against anthrax toxemia by hexa-D-arginine in vitro and in vivo. Infect. Immun. 2004; 72:602–605. doi: 10.1128/IAI.72.1.602-605.2004. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ] -
Schwegmann-We ß els C., Glende J., Ren X., Qu X., Deng H., Enjuanes L., Herrler G. Comparison of vesicular stomatitis virus pseudotyped with the S proteins from a porcine and a human coronavirus. J. Gen. Virol. 2009; 90:1724–1729. doi: 10.1099/vir.0.009704-0. [ DOI ] [ PubMed ] [ Google Scholar ] -
Shirato K., Kawase M., Matsuyama S. Middle East respiratory syndrome coronavirus infection mediated by the transmembrane serine protease TMPRSS2. J. Virol. 2013; 87:12552–12561. doi: 10.1128/JVI.01890-13. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ] -
Shirato K., Kanou K., Kawase M., Matsuyama S. Clinical Isolates of Human Coronavirus 229E Bypass the Endosome for Cell Entry. J. Virol. 2016; 91:91. doi: 10.1128/JVI.01387-16. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ] -
Shulla A., Heald-Sargent T., Subramanya G., Zhao J., Perlman S., Gallagher T. A transmembrane serine protease is linked to the severe acute respiratory syndrome coronavirus receptor and activates virus entry. J. Virol. 2011; 85:873–882. doi: 10.1128/JVI.02062-10. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ] -
Simmons G., Gosalia D.N., Rennekamp A.J., Reeves J.D., Diamond S.L., Bates P. Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry. Proc. Natl. Acad. Sci. USA. 2005; 102:11876–11881. doi: 10.1073/pnas.0505577102. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ] -
Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell. 2020; 181:281–292.e6. doi: 10.1016/j.cell.2020.02.058. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ] -
Wang C., Horby P.W., Hayden F.G., Gao G.F. A novel coronavirus outbreak of global health concern. Lancet. 2020; 395:470–473. doi: 10.1016/S0140-6736(20)30185-9. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ] WHO Novel Coronavirus(2019-nCoV) Situation Report 52. 2020. https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200312-sitrep-52-covid-19.pdf?sfvrsn=e2bfc9c0_4 WHO Coronavirus disease 2019 (COVID-19) Situation Report – 81. 2020. https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200410-sitrep-81-covid-19.pdf?sfvrsn=ca96eb84_2 -
Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020; 367:1260–1263. doi: 10.1126/science.abb2507. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ] -
Yang Y., Du L., Liu C., Wang L., Ma C., Tang J., Baric R.S., Jiang S., Li F. Receptor usage and cell entry of bat coronavirus HKU4 provide insight into bat-to-human transmission of MERS coronavirus. Proc. Natl. Acad. Sci. USA. 2014; 111:12516–12521. doi: 10.1073/pnas.1405889111. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ] -
Yuan Y., Cao D., Zhang Y., Ma J., Qi J., Wang Q., Lu G., Wu Y., Yan J., Shi Y., et al. Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains. Nat. Commun. 2017; 8:15092. doi: 10.1038/ncomms15092. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ] -
Zhang T., Wu Q., Zhang Z. Probable Pangolin Origin of SARS-CoV-2 Associated with the COVID-19 Outbreak. Curr. Biol. 2020; 30:1346–1351.e2. doi: 10.1016/j.cub.2020.03.022. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ] -
Zhou Y., Vedantham P., Lu K., Agudelo J., Carrion R., Jr., Nunneley J.W., Barnard D., P ö hlmann S., McKerrow J.H., Renslo A.R., Simmons G. Protease inhibitors targeting coronavirus and filovirus entry. Antiviral Res. 2015; 116:76–84. doi: 10.1016/j.antiviral.2015.01.011. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ] -
Zhou H., Chen X., Hu T., Li J., Song H., Liu Y., Wang P., Liu D., Yang J., Holmes E.C., et al. A novel bat coronavirus reveals natural insertions at the S1/S2 cleavage site of the Spike protein and a possible recombinant origin of HCoV-19. bioRxiv. 2020 doi: 10.1101/2020.03.02.974139. [ DOI ] [ Google Scholar ] -
Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020; 579:270–273. doi: 10.1038/s41586-020-2012-7. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ] -
Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., et al. China Novel Coronavirus Investigating and Research Team A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020; 382:727–733. doi: 10.1056/NEJMoa2001017. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]